R in Jupyter
Jupyter allows for language kernels other than Python.
R version
There two versions of R on Picotte. Pick one, and set up your .bashrc
file to use that one by default. See the article on R.
For this article, we will use the default R 4.0.2 (without loading a modulefile).
If you want to use R 4.1.3 via the modulefile R/4.1.3, make sure to
add this line to your .bashrc file so that it becomes your default R.
You will also need to do
module load R/4.1.3
before you follow any of the following steps.
Prerequisites
VS Code Jupyter setup
This requires that you have VS Code set up to use remote Jupyter kernels. See: Jupyter via VS Code
We will use the following Python version (Jupyter is a Python application; the Jupyter kernel can be other languages):
[juser@picotte001 ~]$ module load python/gcc/3.10
R Packages
For an R kernel, some R packages need to be installed.
- roxygen2
- devtools
- IRkernel[1][2]
At the R command line (i.e. in a terminal in VS Code just type
"R
[juser@picotte001 ~]$ module load R
[juser@picotte001 ~]$ R
R version 4.2.2 (2022-10-31) -- "Innocent and Trusting"
Copyright (C) 2022 The R Foundation for Statistical Computing
Platform: x86_64-pc-linux-gnu (64-bit)
...
> install.packages("roxygen2")
> install.packages("devtools")
Warning in install.packages("devtools") :
'lib = "/opt/microsoft/ropen/4.0.2/lib64/R/library"' is not writable
Would you like to use a personal library instead? (yes/No/cancel) yes
Would you like to create a personal library
‘~/R/x86_64-pc-linux-gnu-library/4.0’
to install packages into? (yes/No/cancel) yes
... [some minutes of installation and lots of output] ...
> install.packages("IRkernel")
> IRkernel::installspec(name = 'ir42', displayname = 'R 4.2') # modify this if you're using a version of R different from 4.2
Then, exit R by typing "Ctrl-D".
N.B.
install.packages()in R 4.1.3 may prompt you to select a mirror site. Choose one in the US close to Philadelphia.- some of these packages may have already been installed globally, so you may see output that indicates that fact.
- if any package installation fails, wait a few minutes and try again
Jupyter Lab extension
If using python/gcc/3.10, this should already be installed.
Run Jupyter Lab
Next, run Jupyter Lab. In the terminal:
[juser@picotte001 ~]$ jupyter-lab
[I 2022-09-29 16:09:18.115 ServerApp] jupyterlab | extension was successfully linked.
[I 2022-09-29 16:09:18.124 ServerApp] nbclassic | extension was successfully linked.
[I 2022-09-29 16:09:18.776 ServerApp] notebook_shim | extension was successfully linked.
[I 2022-09-29 16:09:18.841 ServerApp] notebook_shim | extension was successfully loaded.
[I 2022-09-29 16:09:18.841 LabApp] JupyterLab extension loaded from /ifs/opt/python/gcc/3.10/lib/python3.10/site-packages/jupyterlab
[I 2022-09-29 16:09:18.841 LabApp] JupyterLab application directory is /ifs/opt/python/gcc/3.10/share/jupyter/lab
[I 2022-09-29 16:09:18.845 ServerApp] jupyterlab | extension was successfully loaded.
[I 2022-09-29 16:09:18.862 ServerApp] nbclassic | extension was successfully loaded.
[I 2022-09-29 16:09:18.862 ServerApp] Serving notebooks from local directory: /home/rbatty
[I 2022-09-29 16:09:18.862 ServerApp] Jupyter Server 1.18.1 is running at:
[I 2022-09-29 16:09:18.862 ServerApp] http://localhost:8888/lab?token=random_characters
...
[I 2022-09-29 16:09:18.862 ServerApp] or http://127.0.0.1:8888/lab?token=random_characters
[I 2022-09-29 16:09:18.862 ServerApp] Use Control-C to stop this server and shut down all kernels (twice to skip confirmation).
[C 2022-09-29 16:09:18.882 ServerApp]
To access the server, open this file in a browser:
file:///home/rbatty/.local/share/jupyter/runtime/jpserver-14372-open.html
Or copy and paste one of these URLs:
http://localhost:8888/lab?token=random_characters
or http://127.0.0.1:8888/lab?token=random_characters
Copy the the URL shown (containing the token of random characters. Open a new browser window on your PC and paste the URL into the address bar.
From Jupyter Lab's "File" menu, select "New Launcher". You should see a new tab, where you can click on the "R" button under "Notebook":

When you click it, you should have a new notebook using the R kernel.
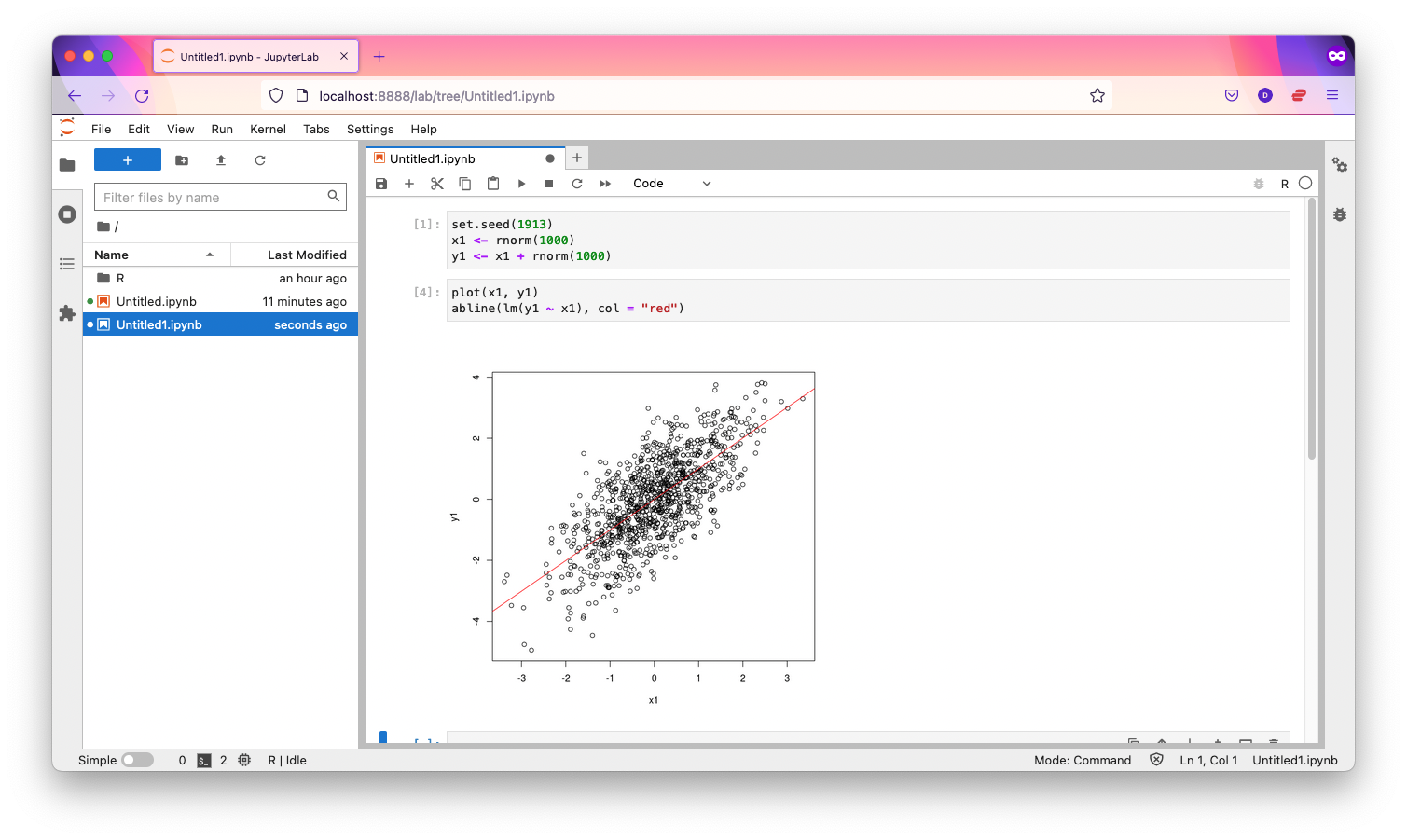
NOTE
- these files reside on Picotte, not on your PC.
- the R application runs on `picotte001`, the login node, which is concurrently used by other users. Please do not run large computations in a notebook like this.
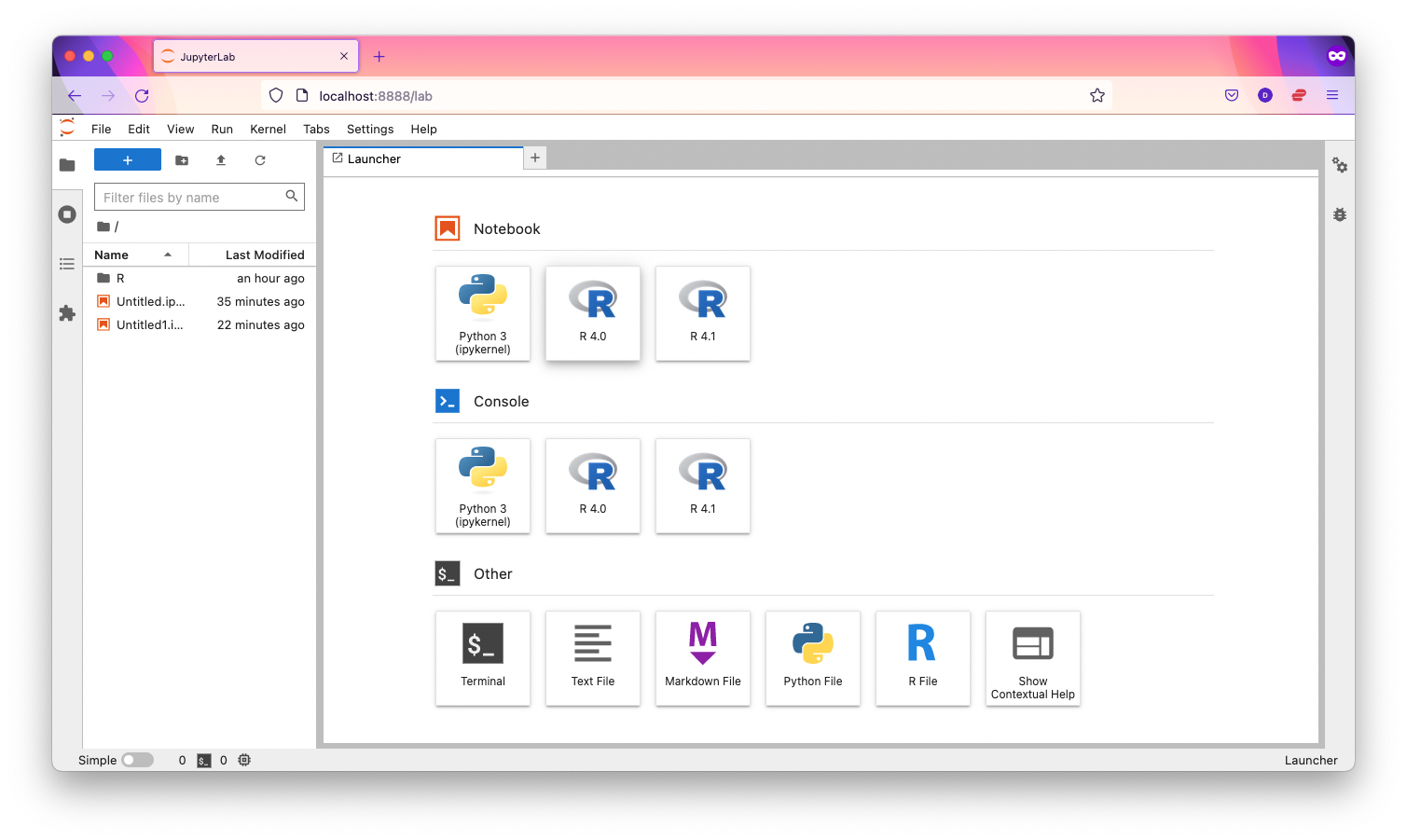
Multiple kernels for multiple versions of R
It is also possible to have separate kernels for R 4.0 and R 4.1:
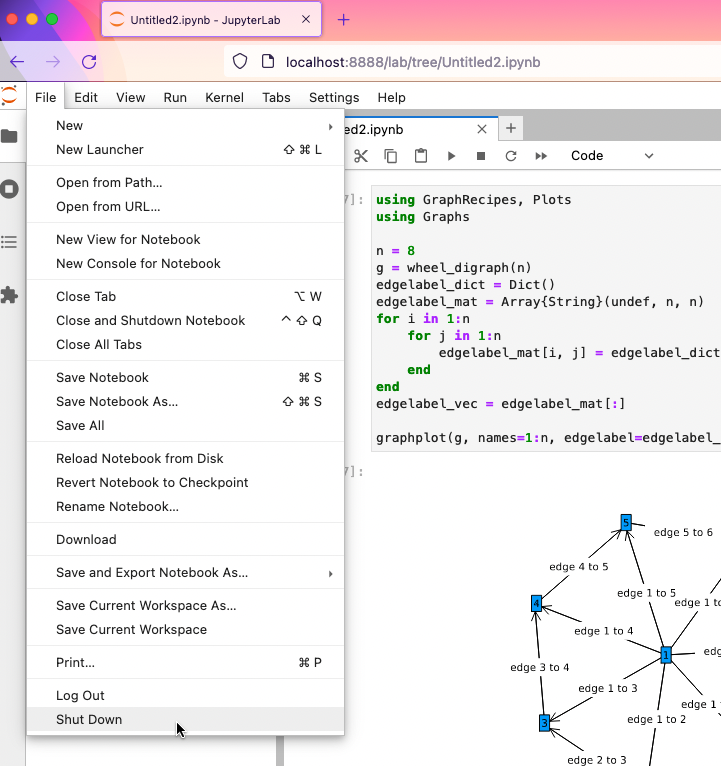
Finishing up
When done working, please select "Shut Down" from the Jupyter Lab "File" menu.
See Also
References
[1] IRkernel website