NAMD
NAMD is a molecular dynamics simulation package.[1]
General Note
- All the ibverbs versions should use the
namdNN("NN" is a number) PEs
Installed Versions
CPU-only
NAMD 2.10 using OpenMPI and Intel MKL. Use the modulefile:
namd/intel/2015/openmpi/2.10
This also loads the following, so do not load the older Intel compiler or MKL modules, or any other MPI modules:
- intel/composerxe/2015.1.133
- proteus-openmpi/intel/2015/1.8.1-mlnx-ofed
The build uses
- FFTW3 from the Intel Math Kernel Library (MKL)
- Mellanox MXM messaging accelerator (via OpenMPI 1.8.1)
Also available is the binary-distributed NAMD-2.12-ibverbs-smp but the job may need to generate a correct host list file.
CPU-only for 40-core Haswell nodes (new.q) using ibverbs
NAMD 2.13 using ibverbs and Intel MKL. Use the modulefile:
namd/intel/2019/ibverbs/2.13
This also loads the following:
intel/composerxe/2019u1hpcx-stack(for Mellanox optimized Infiniband interface)
Use the namd40 PE. Example job script:
#!/bin/bash
#$ -S /bin/bash
#$ -cwd
#$ -j y
#$ -P myrsrchPrj
#$ -M myname@drexel.edu
#$ -pe namd40 80
#$ -l h_rt=02:00:00
#$ -l h_vmem=2G
#$ -l m_mem_free=2G
#$ -q new.q
. /etc/profile.d/modules.sh
module load shared
module load sge/univa
module load proteus
module load proteus-rh68
module load namd/intel/2019/ibverbs/2.13
### NOTE there are currently (2019-03-15) only 3 nodes available for general use.
charmrun ++scalable-start ++p $NSLOTS ++nodelist ./nodelist.${JOB_ID} ${NAMD2EXE} +setcpuaffinity ++verbose apoa1.namd
/bin/rm -f ./nodelist.${JOB_ID}
Benchmarks
The standard apoa1 benchmark was run. Unfortunately, there was other
load on the cluster for some of the following runs, so it is not a clear
indication of performance. Benchmark was run on 32 processors, with the
32 distributed in different ways:
- 2 nodes, 16 procs per node: 32 CPUs 0.0463154 s/step 0.536058 days/ns 626.434 MB memory
- 4 nodes, 8 procs per node (under load from other jobs): 32 CPUs 0.0376342 s/step 0.435581 days/ns 598.762 MB memory
- 8 nodes, 4 procs per node (under load from other jobs): 32 CPUs 0.0605087 s/step 0.700332 days/ns 587.395 MB memory
On the new 40-core Haswell nodes, available in new.q:
- 1 node, 40 procs per node: 40 CPUs 0.0184217 s/step 0.213214 days/ns 491.844 MB memory
- 2 nodes, 40 procs per node: 80 CPUs 0.00956029 s/step 0.110651 days/ns 494.816 MB memory
- 3 nodes, 40 procs per node: 120 CPUs 0.00673581 s/step 0.0779608 days/ns 493.559 MB memory
apoa1 input file
Important: modify the file apoa1.namd[2] so that its output file
does not clash with other users' output.
outputname /usr/tmp/apoa1-out-myname
Example job script for Open MPI versions (NOT ibverbs):
#!/bin/bash
#$ -S /bin/bash
#$ -N testnamd
#$ -cwd
#$ -j y
#$ -P fixmePrj
#$ -M fixme@drexel.edu
#$ -pe fixed16 32
### OR -pe openmpi_ib 32
### if you do not care how the processes are distributed
#$ -l h_rt=00:30:00
#$ -l h_vmem=2G
#$ -l m_mem_free=2G
#$ -l vendor=intel
#$ -q all.q
. /etc/profile.d/modules.sh
module load shared
module load proteus
module load gcc/4.8.1
module load sge/univa
module load namd/intel/2015/openmpi/2.10
which namd2
${MPI_RUN} namd2 apoa1.namd
### For some reason not understood, you may need to have mpirun set the LD_LIBRARY_PATH for all slave processes.
### Do this if you see errors like:
### namd2: /usr/lib64/libstdc++.so.6: version `GLIBCXX_3.4.15' not found (required by namd2)
# mpirun -x LD_LIBRARY_PATH namd2 apoa1.namd
#
# You may also use the full path to mpirun, or use the "--prefix ${MPI_HOME}" argument.
# They are equivalent, and force mpirun to search the ${MPI_HOME} prefix for libraries.
# All 3 below are equivalent:
# mpirun --prefix=${MPI_HOME} namd2 apoa1.namd
# ${MPI_RUN} namd2 apoa1.namd
# ${MPI_HOME}/mpirun namd2 apo1.namd
The output file will be named testnamd.oNNNNNN.
CUDA-enabled Version using MPI
CUDA-enabled builds can run on GPU nodes. CUDA + MPI is not recommended as it may not perform well. See the next section for the recommended method to run CUDA code over multiple nodes.
namd/gcc-cuda/openmpi/2.9
Online documentation: http://www.ks.uiuc.edu/Research/namd/2.12/ug/
Example job script - runs on all 8 GPU nodes. Fewer GPU nodes may be requested by reducing the number of requested slots in steps of 16, e.g. "-pe namdgpu 64" will give 4 GPU nodes.
NB Grid Engine vmem accounting for GPU jobs may be incorrect. If the
job terminates quickly, increase the h_vmem request, even above the
physical limit.
#!/bin/bash
#$ -S /bin/bash
#$ -cwd
#$ -j y
#$ -P urcfadmPrj
#$ -M dwc62@drexel.edu
#$ -pe fixed16 32
#$ -l h_rt=00:30:00
#$ -l h_vmem=200G
#$ -l m_mem_free=2G
#$ -l ngpus=2
#$ -q gpu.q
. /etc/profile
module load shared
module load proteus
module load sge/univa
module load gcc
module load namd/cuda60/openmpi/2.9
### NAMD2EXE is set in the modulefile
${MPI_RUN} ${NAMD2EXE} apoa1.namd
/bin/rm -f FFTW_NAMD*.txt
In the job output, you should see messages listing the GPU (CUDA) devices being used:
Info: 1 NAMD 2.9 Linux-x86_64-MPI-CUDA 32 gpu03 dwc62
Info: Running on 32 processors, 32 nodes, 2 physical nodes.
Info: CPU topology information available.
Info: Charm++/Converse parallel runtime startup completed at 0.03901 s
Pe 17 physical rank 1 will use CUDA device of pe 16
Pe 21 physical rank 5 will use CUDA device of pe 16
Pe 31 physical rank 15 will use CUDA device of pe 24
Pe 16 physical rank 0 binding to CUDA device 0 on gpu05: 'Tesla K20Xm' Mem: 5759MB Rev: 3.5
Pe 18 physical rank 2 will use CUDA device of pe 16
Pe 19 physical rank 3 will use CUDA device of pe 16
Pe 20 physical rank 4 will use CUDA device of pe 16
Pe 23 physical rank 7 will use CUDA device of pe 16
Pe 25 physical rank 9 will use CUDA device of pe 24
Pe 26 physical rank 10 will use CUDA device of pe 24
Pe 30 physical rank 14 will use CUDA device of pe 24
Pe 27 physical rank 11 will use CUDA device of pe 24
Pe 24 physical rank 8 binding to CUDA device 1 on gpu05: 'Tesla K20Xm' Mem: 5759MB Rev: 3.5
Did not find +devices i,j,k,... argument, using all
Pe 2 physical rank 2 will use CUDA device of pe 4
Pe 3 physical rank 3 will use CUDA device of pe 4
Pe 4 physical rank 4 binding to CUDA device 0 on gpu03: 'Tesla K20Xm' Mem: 5759MB Rev: 3.5
Pe 6 physical rank 6 will use CUDA device of pe 4
Pe 7 physical rank 7 will use CUDA device of pe 4
Pe 15 physical rank 15 will use CUDA device of pe 8
Pe 1 physical rank 1 will use CUDA device of pe 4
Pe 0 physical rank 0 will use CUDA device of pe 4
Pe 5 physical rank 5 will use CUDA device of pe 4
Pe 8 physical rank 8 binding to CUDA device 1 on gpu03: 'Tesla K20Xm' Mem: 5759MB Rev: 3.5
Pe 9 physical rank 9 will use CUDA device of pe 8
Pe 11 physical rank 11 will use CUDA device of pe 8
Pe 13 physical rank 13 will use CUDA device of pe 8
Pe 10 physical rank 10 will use CUDA device of pe 8
Pe 12 physical rank 12 will use CUDA device of pe 8
Pe 14 physical rank 14 will use CUDA device of pe 8
CUDA-enabled Version using IBVERBS
This is the recommended way of running NAMD on multiple nodes while utilizing GPUs. Since MPI combined with CUDA gives poor performance, NAMD can use the lower-level InfiniBand (IB) Verbs interface for internode communication. This uses Charm++ (charmrun) to run NAMD.
Use the modulefile:
namd/ibverbs-smp-cuda/2.12
Official documentation for running the IBVERBS version is here: http://www.ks.uiuc.edu/Research/namd/2.12/ug/node83.html
Official documentation for CUDA acceleration is here: http://www.ks.uiuc.edu/Research/namd/2.12/ug/node90.html
Each namd2 thread can use only one GPU. Therefore you will need to run at least one thread for each GPU you want to use. Multiple threads can share a single GPU, usually with an increase in performance.
NVIDIA has recommendations for running this IBVERBS-CUDA version of NAMD, specifying the number of MPI ranks and threads to ensure efficient use of the GPU devices: https://www.nvidia.com/en-us/data-center/gpu-accelerated-applications/namd/ The contents of that page are summarized below.
NAMD has to be run with this command line:
charmrun {charmOpts} namd2 {namdOpts} {inputFile}
{charmOpts}++nodelist {nodeListFile}- multi-node runs require a list of nodes. This file is automatically generated by thenamdgpuPE++p $totalPes- specifies the total number of PE threads. This is the total number of Worker Threads (aka PE threads). We recommend this to be equal to (#TotalCPUCores - #TotalGPUs).++ppn $pesPerProcess- number of PEs per process. We recommend to set this to (#ofCoresPerNode/#ofGPUsPerNode – 1)- This is necessary to free one of the threads per process for
communication. Make sure to specify <
+commapbelow. - Total number of processes is equal to $totalPes/$pesPerProcess
- When using the recommended value for this option, each process will use a single GPU.
- This is necessary to free one of the threads per process for
communication. Make sure to specify <
{namdOpts}- It is recommended to have no more than one process per GPU in the multi-node run. To get more communication threads, it is recommended to launch exactly one process per GPU.
- CPU affinity options (see NAMD user guide):
- '
+setcpuaffinity' in order to keep threads from moving about - '
+pemap #-#' - this maps computational threads to CPU cores - '
+commap #-#' - this sets range for communication threads. Example for dual-socket configuration with 16 cores per socket:+setcpuaffinity +pemap 1-15,17-31 +commap 0,16
- '
- GPU options (see NAMD user guide):
- '
+devices {CUDA IDs}' - optionally specify device IDs to use in NAMD. Devices are numbered sequentially, starting from 0. On Proteus, the CUDA IDs would be "0,1" - If devices are not in socket order it might be useful to set
this option to ensure that sockets use their
directly-attached GPUs, for example, '
+devices 2,3,0,1'
- '
Example job script:
#!/bin/bash -l
#$ -S /bin/bash
#$ -cwd
#$ -j y
#$ -P FIXME
#$ -M FIXME@drexel.edu
#$ -q gpu.q
#$ -pe namdgpu 32
#$ -l h_rt=00:30:00
#$ -l h_vmem=100G
#$ -l m_mem_free=3G
### gpu is a per-host value
#$ -l gpu=2
. /etc/profile.d/modules.sh
module load shared
module load proteus
module load sge/univa
module load gcc/4.8.1
module load namd/ibverbs-smp-cuda/2.12
### nodelist is generated automatically by the namdgpu PE
NODELIST=./nodelist.${JOB_ID}
INPUTFILE=production_li_1_test.conf
### the "expr" program used below performs arithmetic
### NVIDIA recommendation
# total no. of CPU cores for this job is 32
# total no. of GPUs is 4
TOTALPES=$( expr $NSLOTS - $NHOSTS \* 2 )
# no. of CPU cores per node is 16
# no. of GPUs per node is 2
PESPERPROCESS=$( expr 16 / 2 - 1 )
# CPU affinity options: 2 sockets, 8 cores per socket
# use only the first 7 cores in each socket
AFFINITYOPTIONS="+setcpuaffinity +pemap 1-7,9-15 +commap 0,8"
# OPTIONAL GPU device IDs need not be specified since the default is to use all devices
#GPUOPTIONS="+devices 0,1"
charmrun ++nodelist ${NODELIST} ++p ${TOTALPES} ++ppn ${PESPERPROCESS} ${NAMD2EXE} ${AFFINITYOPTIONS} ${INPUTFILE}
And the job output should show the hardware binding information:
Charmrun> scalable start enabled.
Charmrun> IBVERBS version of charmrun
Charmrun> started all node programs in 2.339 seconds.
Converse/Charm++ Commit ID: v6.7.1-0-gbdf6a1b-namd-charm-6.7.1-build-2016-Nov-07-136676
Charm++> scheduler running in netpoll mode.
CharmLB> Load balancer assumes all CPUs are same.
Charm++> cpu affinity enabled.
Charm++> cpuaffinity PE-core map : 1-7,9-15
Charm++> set comm 0 on node 0 to core #0
Charm++> Running on 2 unique compute nodes (16-way SMP).
Charm++> cpu topology info is gathered in 0.002 seconds.
Info: Built with CUDA version 6050
Did not find +devices i,j,k,... argument, using all
Pe 8 physical rank 8 will use CUDA device of pe 7
Pe 25 physical rank 11 will use CUDA device of pe 21
Pe 26 physical rank 12 will use CUDA device of pe 21
Pe 17 physical rank 3 will use CUDA device of pe 14
Pe 11 physical rank 11 will use CUDA device of pe 7
Pe 22 physical rank 8 will use CUDA device of pe 21
Pe 5 physical rank 5 will use CUDA device of pe 4
Pe 13 physical rank 13 will use CUDA device of pe 7
Pe 16 physical rank 2 will use CUDA device of pe 14
Pe 27 physical rank 13 will use CUDA device of pe 21
Pe 3 physical rank 3 will use CUDA device of pe 4
Pe 19 physical rank 5 will use CUDA device of pe 14
Pe 12 physical rank 12 will use CUDA device of pe 7
Pe 24 physical rank 10 will use CUDA device of pe 21
Pe 18 physical rank 4 will use CUDA device of pe 14
Pe 10 physical rank 10 will use CUDA device of pe 7
Pe 6 physical rank 6 will use CUDA device of pe 4
Pe 23 physical rank 9 will use CUDA device of pe 21
Pe 15 physical rank 1 will use CUDA device of pe 14
Pe 2 physical rank 2 will use CUDA device of pe 4
Pe 9 physical rank 9 will use CUDA device of pe 7
Pe 20 physical rank 6 will use CUDA device of pe 14
Pe 0 physical rank 0 will use CUDA device of pe 4
Pe 1 physical rank 1 will use CUDA device of pe 4
Pe 21 physical rank 7 binding to CUDA device 1 on gpu05: 'Tesla K20Xm' Mem: 5759MB Rev: 3.5
Pe 14 physical rank 0 binding to CUDA device 0 on gpu05: 'Tesla K20Xm' Mem: 5759MB Rev: 3.5
Pe 7 physical rank 7 binding to CUDA device 1 on gpu02: 'Tesla K20Xm' Mem: 5759MB Rev: 3.5
Info: NAMD 2.12 for Linux-x86_64-ibverbs-smp-CUDA
Info:
Info: Please visit http://www.ks.uiuc.edu/Research/namd/
Info: for updates, documentation, and support information.
Info:
Info: Please cite Phillips et al., J. Comp. Chem. 26:1781-1802 (2005)
Info: in all publications reporting results obtained with NAMD.
Info:
Info: Based on Charm++/Converse 60701 for net-linux-x86_64-ibverbs-smp-iccstatic
Pe 4 physical rank 4 binding to CUDA device 0 on gpu02: 'Tesla K20Xm' Mem: 5759MB Rev: 3.5
Info: Built Wed Dec 21 11:42:39 CST 2016 by jim on harare.ks.uiuc.edu
Info: 1 NAMD 2.12 Linux-x86_64-ibverbs-smp-CUDA 28 gpu02 myname
Info: Running on 28 processors, 4 nodes, 2 physical nodes.
Info: CPU topology information available.
Info: Charm++/Converse parallel runtime startup completed at 0.051353 s
CkLoopLib is used in SMP with a simple dynamic scheduling (converse-level notification) but not using node-level queue
...
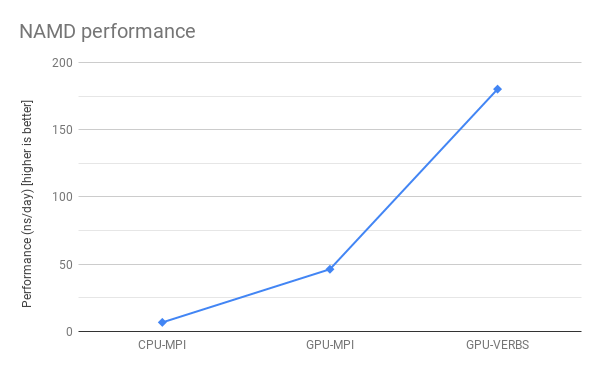
Performance Comparison
A nonrigorous comparison of the same simulation (apoa1.namd) shows the GPU+IBVERBS version runs about 26 times faster than the MPI CPU-only version.
| NAMD flavor | Performance in day/ns (smaller is better) |
|---|---|
| CPU-only 2 nodes (32 slots) MPI Sandy Bridge (using Intel compilers + MKL) | 0.148 |
| CUDA-enabled (4 GPU devices) 2 nodes MPI | 0.0216 |
| CUDA-enabled (4 GPU devices) 2 nodes IB verbs | 0.00555 |
Compiling NAMD
See the Compiling NAMD article.